CASCADE-P Tutorial for http://greengenes.llnl.gov/16S
The CASCADE-P tools use the prokMSA as their data source. The prokMSA is a multiple sequence alignment of the publicly available 16S rDNA sequences. Some functions require the user to indicate which branch(es) of the phylogenetic tree they wish to examine. The “Browse” function allows the user to find and select branches of interest to include in “My Interest List”. To extract an alignment “Slice” or to create a sequence “Consensus” or to search for “Probes”, “My Interest List” must contain at least one phylogenetic branch.
How to BROWSE the sequences in the phylogenetic tree.
The “Browse” tool allows the user to view the categorization of sequences within the prokMSA. The current tree used by CASCADE-P is that proposed by the Ribosomal Database Project v8.1 (http://rdp.cme.msu.edu/). The “Browse” tool is also used to add sequences of interest to “My Interest List” displayed on the left side of the window. Use a recent version of Netscape (ver.6) or Internet Explorer (ver. 5) for full functionality.
1. Beneath the “Functions” menu, select the “Browse” link.
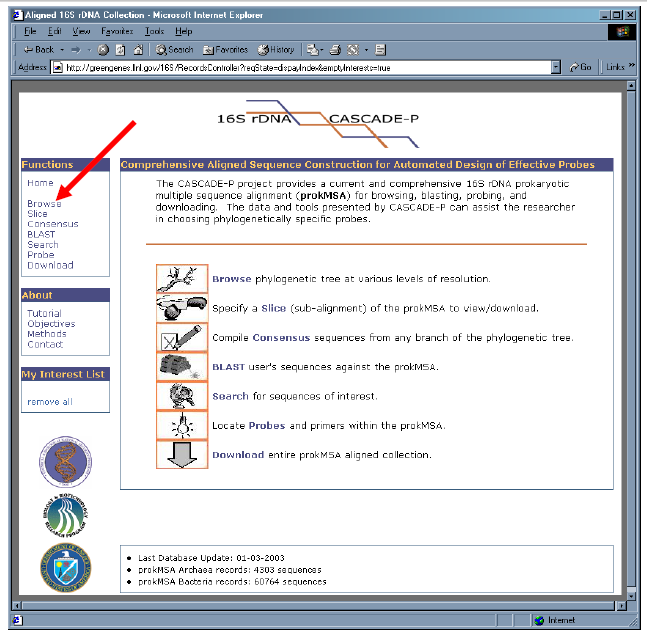
2. Notice that the Browse menu displays the two main branches of the prokaryotic tree. The numbers at left represent the phylocode of each branch. The numbers in parenthesis indicates the quantity of sequences within each branch.
3. Click on “2 – BACTERIA” to reveal its sub-branches.
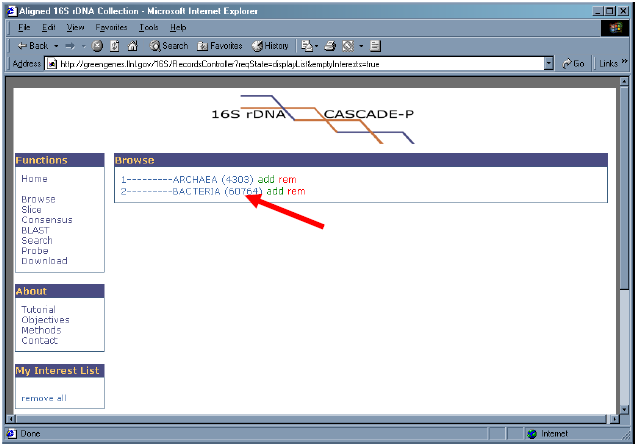
4. Click on “2.2 – THERMOTOGALES” to reveal its sub-branches.
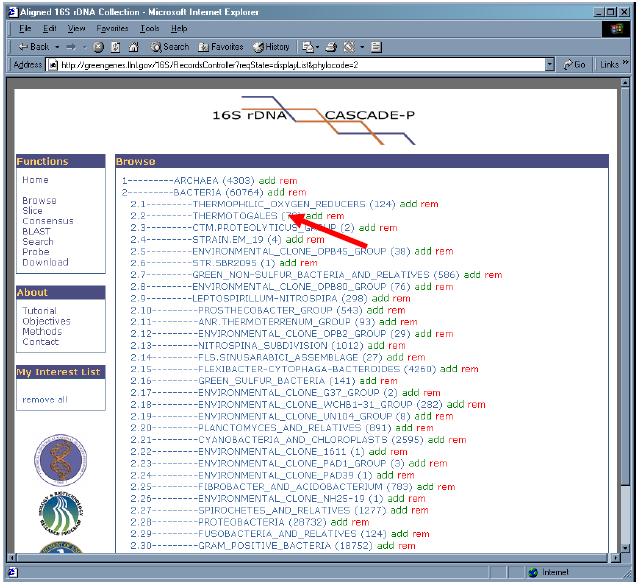
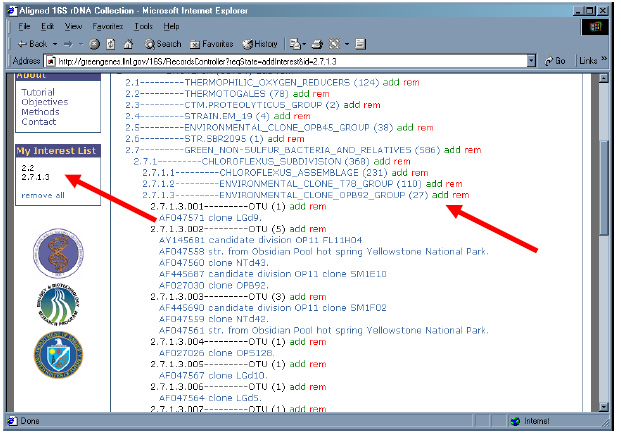
5. Let’s “add” all sequences from phylobranch 2.2 into “My Interest List” by clicking upon “add” to the right of “2.2 – THERMOTOGALES”.
6. Notice that “My Interest List” now contains “2.2”. Which means all sub-branches of 2.2 (2.2.1, 2.2.2, 2.2.3, 2.2.4, 2.2.5, 2.2.6, and 2.2.7) collectively in “My Interest List”.
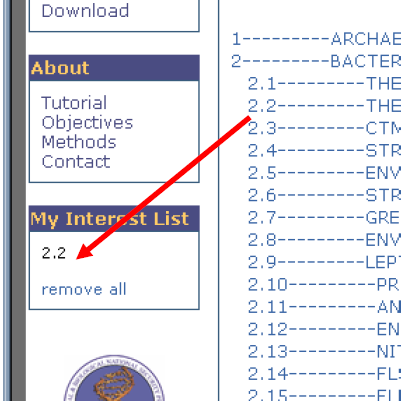
7. In the same manner, now select 2.7, then 2.7.1, then 2.7.1.3 to reveal the OTUs (Operational Taxonomic Units) of the “ENVIRONMENTAL_CLONE-OPB92_GROUP”.
8. Add the sequences of branch 2.7.1.3 by clicking “add” on the 2.7.1.3 line.
9. Notice that 2.7.1.3 now appears in “My Interest List”.
How to extract a Slice (sub-alignment) from the prokMSA.
Note: An Interest List must be established before using the Slice, Consensus or Probe features.
A “Slice” or ‘Sub-alignment” is a portion of the prokMSA which covers a user specified region of the 16S gene. The prokMSA follows the RDP v8.1 convention of stretching each sequence record to 4,182 characters. The default span is from positions 68 to 3,689 a common region targeted by universal primers (Dojka et al. 1998, AEM v64:10, pp3869-77). The user can indicate the beginning and end-points of the desired slice relative to the prokMSA aligned format or relative to the unaligned 16S rDNA format (using standard E. coli numbering). The non aligned E. coli alignment is very approximate, that is, not all 16S rDNA sequences are similar to E. coli. The user can further restrict the sequences that are included based on length and ambiguity thresholds. Formatting options include removing common gap characters which would result in outputted sequences having equal character length. Alternatively, all gap characters can be removed resulting in an unequal alignment output. This type of format may be useful if wanting to create an unaligned, multiple sequence fasta file.
- Beneath the “Functions” menu, select of the “Slice” link.
- Under “Enter character positions relative to aligned format”, enter 2,500 and 3,500 for 5′ and 3′ respectively.
- Under “Select length threshold”, enter 200.
- Under “Select ambiguity threshold”, enter 0.5.
- Under “Select formatting options”, select “remove common alignment gap characters”.
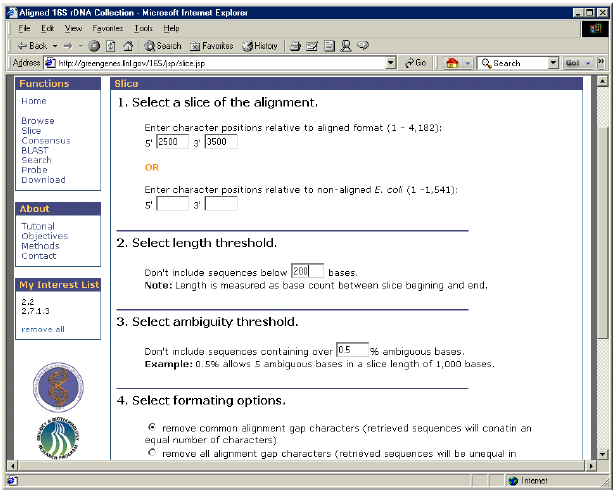
6. After all parameters are chosen, click the “Submit” button. This will create the slice from the sequences in “My Interest List”.
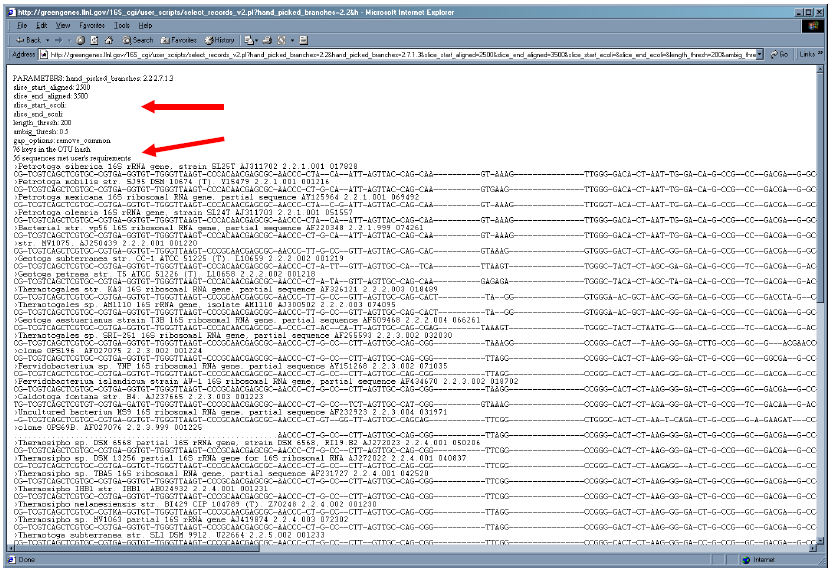
7. Notice that output displays the chosen parameters and reports the number of sequences which met the user’s requirements. The user can scroll to view the entire slice.
8. For each record, the description, GenBank accession number, prokMSA OTU and the prokMSA_id number is shown. The character “.” indicates that no sequence data is available for this position and “-” is an alignment gap character.

9. Save the slice as text by selecting “Save” under the “File” menu of the browser window.
How to create a Consensus from the prokMSA.
Note: An Interest List must be established before using the Slice, Consensus or Probe features.
A “Consensus” is a summary of the nucleotide sequences of prokMSA which covers a user specified region of the 16S rDNA gene. The prokMSA follows the RDP v8.1 convention of stretching each sequence record to 4,182 characters. The default span is from positions 68 to 3,689 a common region targeted by universal primers (Dojka et al. 1998, AEM v64:10, pp3869-77). The user can indicate the beginning and end-points of the desired consensus relative to the prokMSA aligned format or relative to the unaligned 16S rDNA format (using standard E. coli numbering). The non aligned E. coli alignment is very approximate, that is, not all 16S rDNA sequences are similar to E. coli. The user can further restrict the sequences which contribute to the consensus based on length and ambiguity thresholds
- Beneath the “Functions” menu, select of the “Consensus” link.
- Under “Enter character positions relative to aligned format”, enter 2,500 and 3,500 for 5′ and 3′ respectively.
- Under “Select length threshold”, enter 200.
- Under “Select ambiguity threshold”, enter 0.5.
- Under “Select consensus threshold”, enter 70.
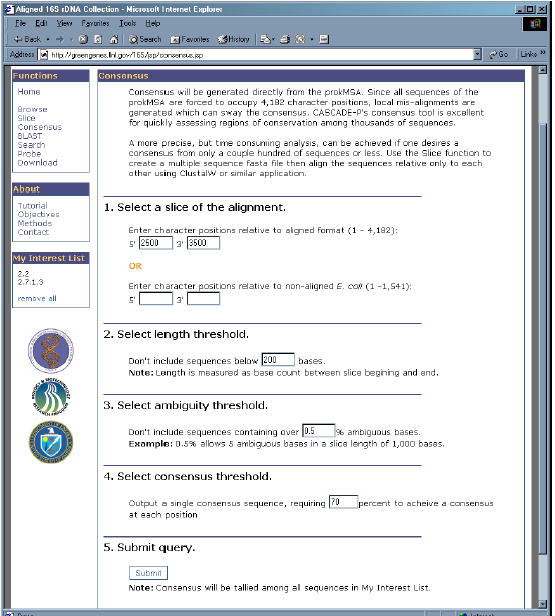
6. After all parameters are chosen, click the “Submit” button. This will create the consensus from the sequences in “My Interest List”.
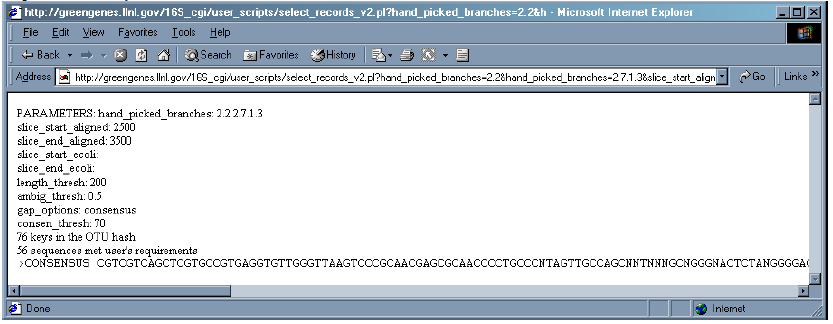
7. Notice that output displays the chosen parameters and reports the number of sequences which met the user’s requirements. The user can scroll to view entire consensus sequence.
8. The character “N” indicates that there was insufficient agreement (below 70% in this case) within the sequences to warrant a consensus base.
9. Save the consensus as text by selecting “Save” under the “File” menu of the browser window or copy and paste it into a text editor.
More info about the prokMSA Consensus function:
Consensus will be generated directly from the prokMSA. Since all sequences of the prokMSA are forced to occupy 4,182 character positions, local mis-alignments are generated which can sway the consensus. CASCADE-P’s consensus tool is excellent for quickly assessing regions of conservation among thousands of sequences.
A more precise, but time consuming analysis, can be achieved if one desires a consensus from only a couple hundred of sequences or less. Use the Slice function to create a multiple sequence fasta file, then align the sequences relative only to each other using ClustalW or a similar application.
How to use the BLAST tool.
Cascade –P offers a BLAST interface to query the prokMSA with a user provided sequence. The sequences can be pasted in fasta or raw text format. There are three collections to compare the sequence query against:
(1) The Ribosomal Database Project (RDP) aligned collection (16,277 sequences in version 8.1).
(2) The complete 16S collection – the entire prokMSA database (which includes RDP v8.1) containing over 60,000 sequences.
(3) The “ highest quality” from the prokMSA database. This only reports sequences which exceed 600 base pairs with less than 0.5% ambiguity (bases that cannot be classified as A, G C or T). Approximately half of the prokMSA is considered “highest quality”.
1. Beneath the “Functions” menu, select of the “BLAST” link.
2. Copy and paste this sequence into the window labeled “paste sequence here:”
>Lab Isolate 4G12-9p
CACGCCGTAAACGATGAGTGCTAAGTGTTGGAGGGTTTCCGCCCTTCAGTGCTGCAGCTAAC
CATTAAGCACTCCGCCTGGGGAGTACGGCCGCAAGGCTGAAACTCAAAGGAATTGACGGGGG
CGCCAAGCGGTGGAGCATGTGGTTTAATTCGAAGCAACGCGAAGAACCTTACCAGGTCTTGA
CATCCCGCTGACCGGTCTGGAGACAGGCCTTTCTTCGGGGACAGCGGTGACAGGTGGTGCAT
GTTGTCGTCAGCTCGTGTCGGAAGGGTTAAGTCCCGCAACGAGCGCAACCCTTGATCAAAGT
TTAGTCCCAGCATTCAGTTGGGCACTCTAAGGTGACTGCCGGTGACAAACCGGAGGAAGGTG
GGGATGACGTCAAATCATCATGCCCCTTATGACCTGGGCTACACACGTGCTACAATGGATGG
ACAAAGGGCTGCAAGACCGGAGGTTTAGCCAATCCCATAAAACCATGCTCAGTTCGGATTGC
AGGCTGCAACTCGCCTGCATGAAGCCGGAATCGCTAGTAATCGCGGATCAGCATGCCGCGGT
AATACGTTCCCGGGCCTTGTACACACCGCCCGTCACAAAACGAGAGTTTGCAACACCCGAAG
CGGTGGGGTAACCTTACGGGAGCCA
3. Click “Submit” to accept the default options and to begin the BLAST search.
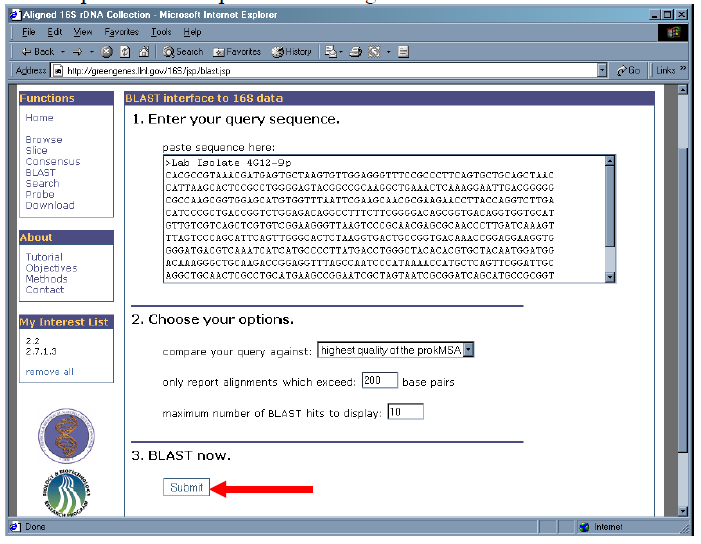
4. Results are returned as a ”phylotree” and display the taxonomy of the best BLAST hits. The descriptions of the most similar sequences to the query are listed within their OTU. The BLAST scores are also listed.
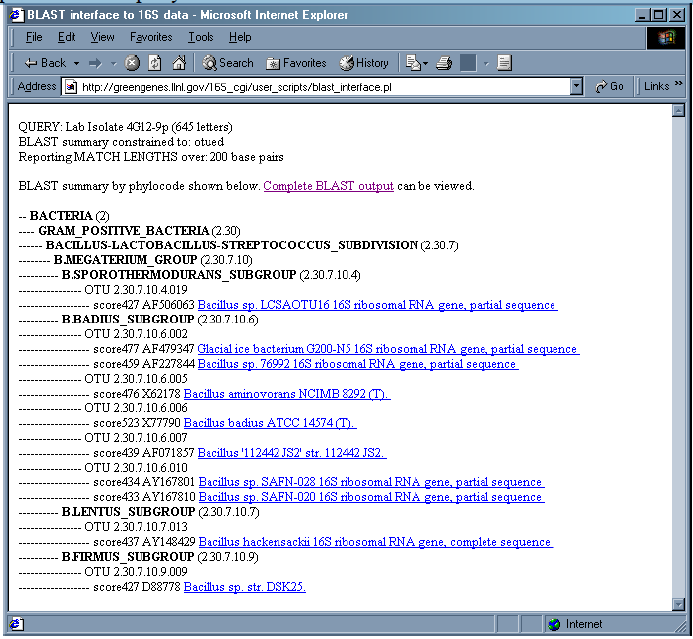
5. The “Complete BLAST output” is also available for viewing/saving.
How to search the 16S rDNA records of the prokMSA
Use this tool to search for a descriptive word identification number. This tool does not search through the sequence data. It basically allows a search of everything in a record apart from the sequence data. If the user wants to search for a specific short sequence use the Probe tool. To search for a long sequence, use BLAST.
There are three collections that can be searched:
(1) The Ribosomal Database Project (RDP) aligned collection (16,277 sequences in version 8.1).
(2) The complete 16S collection – the entire prokMSA database (which includes RDP v8.1) containing over 60,000 sequences.
(3) The “ highest quality” from the prokMSA database. This only reports sequences which exceed 600 base pairs with less than 0.5% ambiguity (bases that cannot be classified as A, G C or T). Approximately half of the prokMSA is considered “highest quality”.
There are multiple data fields that can be searched:
(1) prokMSA identifier – unique integer assigned to each record in the prokMSA collection.
(2) NCBI accession number – alpha numeric value assigned by NCBI.
(3) NCBI unique identifier – unique integer assigned by NCBI.
(4) RDP identifier – alpha numeric value assigned by RDP for some sequences.
(5) Sequence description – annotation assigned by the researcher.
(6) Phylogenetic numeric code – Hierarchal taxonomic designation (Example: 2.30.7.12)
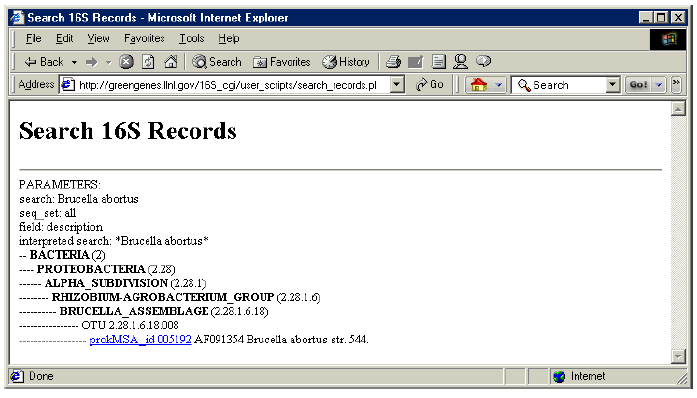
1. In the “Enter search string” box, type “Brucella abortus”.
2. Under “Choose your options” choose “complete prokMSA” for “data set” and “Sequence description” for “data field”.
3. Click on “Submit”.
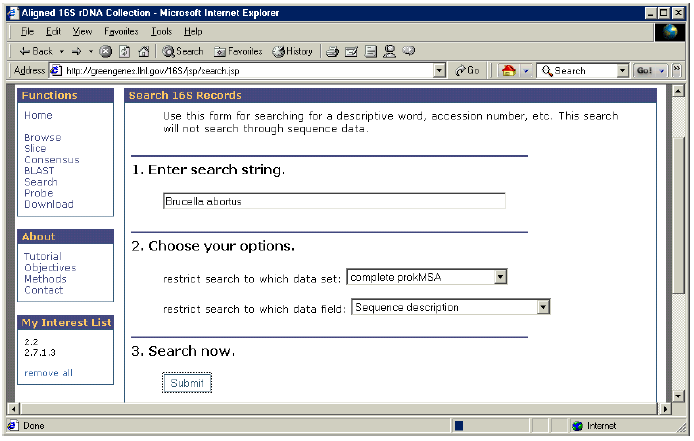
4. Notice that results are presented with phylogenetic information.
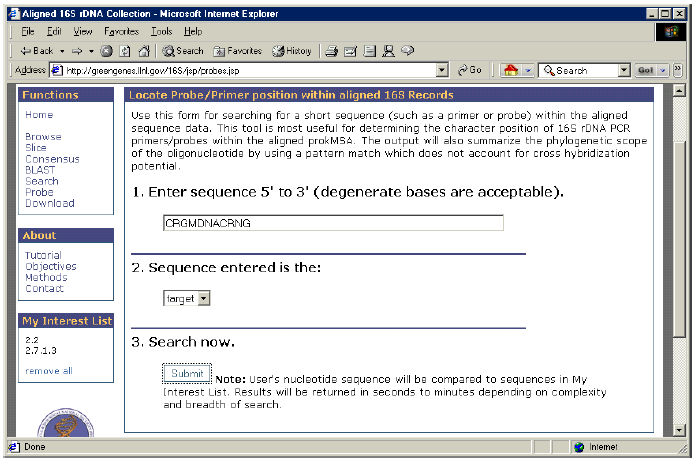
How to locate Probes and Primers within the aligned sequence data.
Note: An Interest List must be established before using the Slice, Consensus or Probe features.
The Probe function can be used to locate short sequences within the aligned sequences of the prokMSA. It is useful for determining the locus of a particular oligomer in relation to the 4,182-character alignment format. Enter sequence (degenerate bases are acceptable) in the 5′-3’ direction and specify whether the entered sequence is the target or the probe. The output data will display the regular expression search pattern used by Perl’s pattern match. Also, the output will specify how many sequences match the query sequence out of the total number of sequences searched. The mean position of matches will be displayed with the sample’s standard deviation which indicates how much variation was encountered in the loci of the matches. The measurements are reported relative to the 4,182-character aligned stretch.
1. Enter the degenerate sequence “CRGMDNACRNG”.
2. Under “Sequence entered is the”, select “target”.
3. Click on “Submit”.
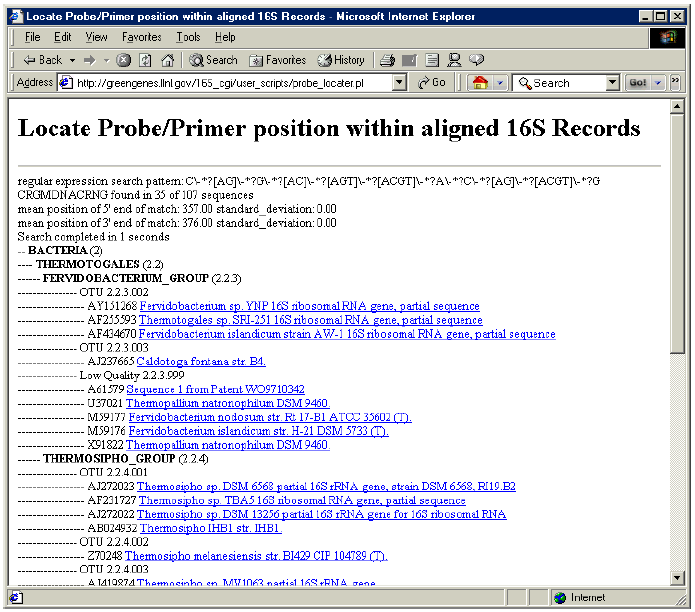
4. The results display the frequency and the location of the pattern match within the sequences represented by “My Interest List”. The matching sequences are listed with phylogenetic information.
Glossary
Acronyms:
CASCADE-P- Comprehensive Aligned Sequence Construction for Automated Design of Effective Probes
prokMSA Prokaryotic Multiple Sequence Alignment
BLAST Basic Local Alignment Search Tool
RDP Ribosomal Database Project
OTU Operational Taxonomic Unit.
16S – the gene encoding 16S ribosomal RNA, an integral part of the small subunit (SSU) of the ribosome.
Align0 – A publicly available sequence aligner distributed with FASTAv2.0u66 (Pearson et al., 1988). Useful for pair-wise global alignment of two sequences while preserving existing hyphen gap characters.
BLAST (Basic Local Alignment Search Tool). CASCADE –P offers a BLAST interface to query the prokMSA with unknown sequences.
CASCADE-P Comprehensive Aligned Sequence Construction for Automated Design of Effective Probes. The CASCADE-P project offers a comprehensive alignment database (prokMSA) of 16S rDNA which enables the design of effective probes.
CLUSTAL W- A Multiple Sequence Alignment program capable of generating dendograms.
Consensus Sequence– A linear series of nucleotides, commonly with gaps and some degeneracy, that define common features of homologous sequences or recognition sites for proteins that act on or bind to nucleic acids.
Eukaryotes – Organisms with nuclei
Hybridization- The reaction by which the pairing of complementary strands of nucleic acid occurs. DNA is usually double-stranded, and when the strands are separated they will re-hybridize under the appropriate conditions. Hybrids can form between DNA-DNA, DNA-RNA or RNA-RNA. They can form between a short strand and a long strand containing a region complementary to the short one. Imperfect hybrids can also form, but the more imperfect they are, the less stable they will be (and the less likely to form). To “anneal” two strands is the same as to “hybridize” them.
Microarray Base-pairing (i.e., A-T and G-C for DNA; A-U and G-C for RNA) or hybridization is the underlining principle in the design of a DNA microarray. An array is an orderly arrangement of samples. It provides a medium for matching known and unknown DNA samples based on base-pairing rules and automating the process of identifying the unknowns. An array experiment can make use of common assay systems such as microplates or standard blotting membranes, and can be created by hand or make use of robotics to deposit the sample. High density microarrays from Affymetrix (Santa Clara, CA, USA) are manufactured using microlithography and can contain 500, 000 probes.
NAST – Nearest Alignment Space Termination. An algorithm which strategically removes gap characters from a pair-wise sequence alignment to compress the alignment to a certain number of characters. NAST produces some local misalignments to force the resulting sequence pair to occupy a fixed number of characters. This tool allows newly discovered sequences to be merged into any existing MSA providing the trade-off between fixed total alignment string length and the extent of local misalignment is acceptable.
Probe A fragment of DNA or RNA which is labeled in some way (often incorporating 32P or 35S or a fluorescent molecule- used for detection) and is used to hybridize with the nucleic acid in which you are interested.
Prokaryotes – Organisms lacking nuclei
Ribosome– A complex ribonucleoprotein particle which translates mRNA into protein molecules
Glossary of CASCADE-P Tutorial – Download [Optimized PDF]
Glossary of CASCADE-P Tutorial – Download